- About
-
Solutions
-
Services
- Biosciences
- Chemistry
- Integrated Drug Discovery
- Computer Aided Drug Design
- Hit Identification
- Target Classes and Modalities
- Therapeutic Areas
-
A-Z
- A
- B
- C
- D
- E
- F
- G
- H
- I
- K
- L
- M
- N
- O
- P
- R
- S
- T
- V
- X
-
Services
- Library
- News & Events
- Careers
Targeted Protein Degradation
There is currently great drug discovery interest and investment in the field of targeted protein degraders as a means of tackling undruggable protein targets.1,2 The catalytic degrader mode-of-action (MOA) offers several potential advantages over conventional small molecule inhibitors. These include prolonged pharmacodynamic (PD) effects and pharmacokinetic (PK)/PD disconnects, facilitating intermittent and reduced dosing regimens, while simultaneously reducing the potential for toxicity. Furthermore, because degrader pharmacology is derived from an event driven process, target ligands are not restricted to catalytic binding pockets. Resulting protein target destruction also removes any residual scaffolding and non-catalytic functions a target possesses, which are usually inaccessible to small molecule inhibitors.
Proteolysis targeting chimeras (PROTACs®) are one type of protein targeted degrader. These heterobifunctional molecules consist of a linker and two ligands, one binding a target protein and the other to an E3 ubiquitin ligase. Subsequent efficient ternary complex formation can result in polyubiquitination of the target protein and degradation via the ubiquitin-proteasome pathway. The field has gained significant momentum, with several agents in advanced clinical trials with an emphasis on multiple cancer indications. However, non-oncology therapeutic areas, such as inflammation and CNS, are beginning to attract attention.
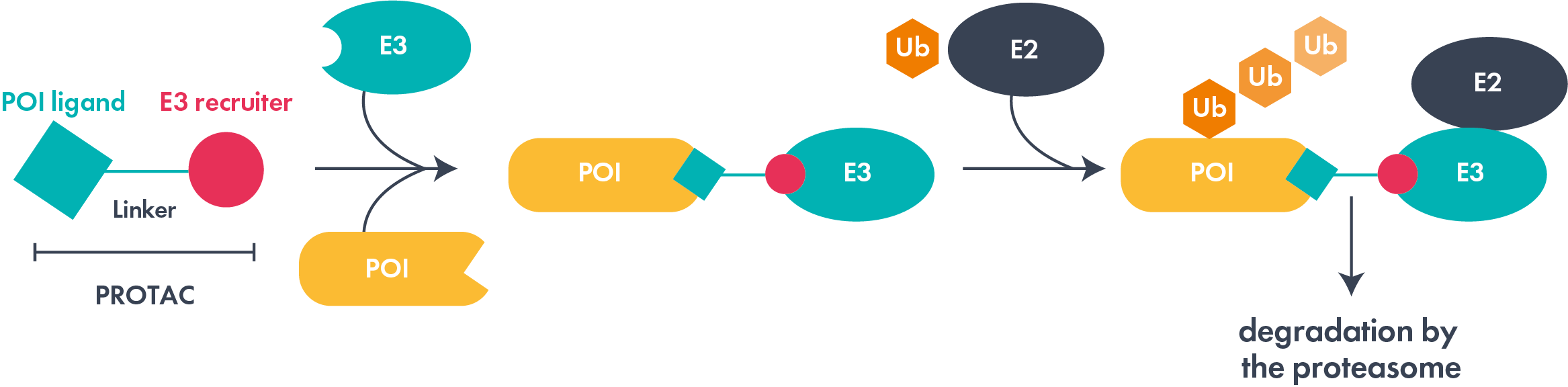
Molecular glues can also act as targeted degraders; find out more on our dedicated molecular glue page. Much earlier in their evolution, lysosome targeting chimeras (LYTACs) and autophagy targeted chimeras (AUTACs) represent exciting areas of protein degradation exploration and research.
Targeted Protein Degradation Research at Domainex
Domainex can provide a fully integrated team to work on your programme with expertise in the design, synthesis and profiling of targeted heterobifunctional protein degraders. Our protein scientists are also able to produce and characterize proteins to support assay development and compound profiling.
Ligand Identification
Domainex can review the literature in order to identify suitable ligands for your target protein of interest. Alternatively if no appropriate ligands are available, Domainex has a range of hit identification platforms, structural biology and computational chemistry capabilities which can be used to rapidly identify ligands with desirable properties and with suitable vectors for linker attachment, which can be used as starting points for your programme. Domainex can provide guidance on the most suitable approach for your programme, taking into consideration the science and any budgetary or timeline constraints.
Domainex has experience discovering E3 ligase ligands and can utilize non-covalent and covalent screening methods to enable the discovery of the next-generation of degraders. For an example of covalent screening methods, see our poster entitled “Covalent Fragment Screening Using QToF Analysis.”
Design and synthesis of degraders
Our talented computational chemists are able to design virtual degrader libraries which can be synthesized by our skilled synthetic chemists who have experience synthesizing degraders incorporating Cereblon, Von Hippel–Lindau tumour suppressor (VHL) and Inhibitor of apoptosis (c/XIAP) E3 ligase ligands at up to gram scale.
As your project progresses, our scientists are able to design modifications to improve the degradation efficiency. As part of this optimization process they will consider the ligand, the vectors of the linker and the linker length and type, while monitoring the physicochemical and ADME properties from an early stage to ensure suitable pharmacokinetic properties are achieved.
Profiling of degraders
Domainex is experienced at establishing in vitro binary and ternary complex formation assays to confirm target engagement using biophysical techniques (Grating Coupled Interferometry (GCI), Spectral Shift or MicroScale Thermophoresis (MST)) and AlphaScreen. Further assays include the use of nanoBRET technology to confirm intracellular target engagement and confirmation of cellular protein degradation using Promega HiBit, western blots and in vitro ubiquitination based assays to characterize each stage of the degrader mode-of-action.
If you would like to access Domainex’s targeted protein degrader expertise to support your own research programme we would be delighted to hear from you.
Direct-to-Biology
Domainex can apply a Direct-to-Biology (D2B) approach to your TPD project. D2B combines plate-based chemistry with rapid screening, synergistically enhancing the efficiency and success of drug discovery. Find out more on our dedicated D2B webpage.
Additionally, we have generated an in-house ‘toolbox’ of partial degraders available for immediate reaction with protein of interest (POI) binders which can be combined with our D2B platform to further accelerate SAR generation.
References
- Resolute Research Report (2021). Targeted protein degradation market landscape https://blog.resolute.ai/resolute-research-reports/targeted-protein-degradation?hs_amp=true
- The PROTAC gold rush. Garber, K. Nature Biotechnology 40, 12-16, 2022
Domainex press release in the protein degradation field:
PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license.
Download the Brochure
Accelerate your targeted protein degrader programmes with access to our toolbox of PROTACs. Download the brochure to find out more.
Start your next project with Domainex
Contact one of our experts today