- About
-
Solutions
-
Services
- Biosciences
- Chemistry
- Integrated Drug Discovery
- Computer Aided Drug Design
- Hit Identification
- Target Classes and Modalities
- Therapeutic Areas
-
A-Z
- A
- B
- C
- D
- E
- F
- G
- H
- I
- K
- L
- M
- N
- O
- P
- R
- S
- T
- V
- X
-
Services
- Library
- News & Events
- Careers
Dynamic Light Scattering (DLS)
Monitoring protein oligomeric state and aggregation.

In Dynamic Light Scattering (DLS), light from a laser is fired at the sample and the scattering pattern of the light is measured. The scattering pattern is closely related to the size and distribution of the particles in solution.
Dynamic light scattering services at Domainex

Domainex has invested in a SpectroLight 600 from Xtal Concepts. The Spectrolight 600 allows us to measure the monodispersity of a protein sample, test for aggregation and more. For example, the quantitative data produced enables efficient stability analyses and buffer screens. No sample preparation is required for DLS and it is quick and easy to measure a sample.
DLS data often provides a good indication of the quality of a sample and its suitability for downstream applications such as X-ray crystallography. It allows us to probe particle size, and therefore detect aggregation as well as monitor the oligomeric state of a protein. Recently this has been applied to proteins solubilised using our PoLiPa technology – allowing us to monitor the effectiveness of PoLiPa solubilization of a membrane protein.
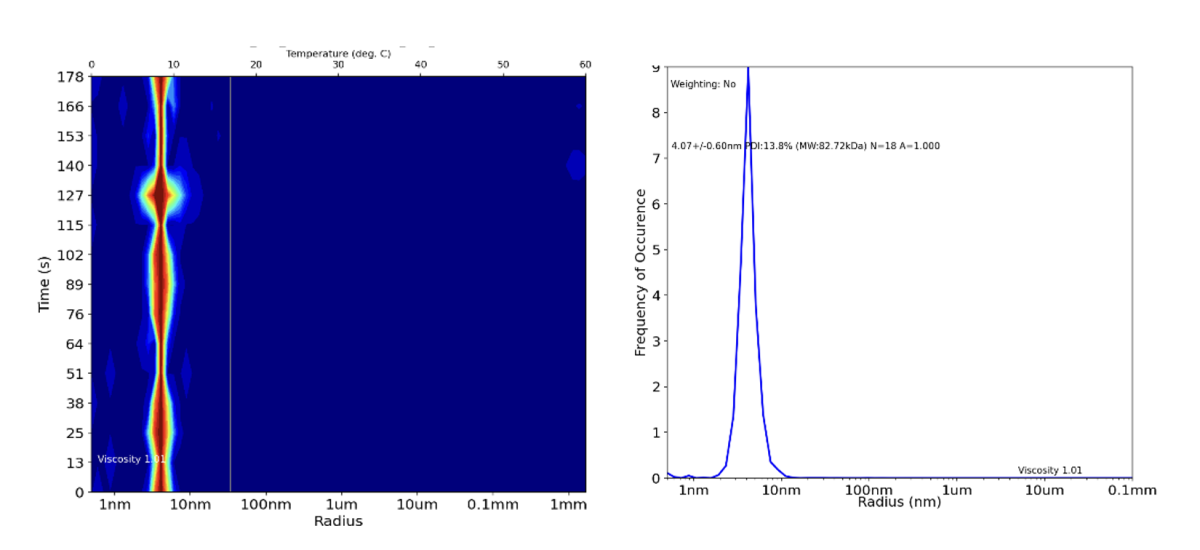
Figure 1: DLS data output for pure soluble protein. This result indicates that the sample is dimeric protein which shows no signs of aggregation over time and is therefore suitable for downstream applications such as X-ray crystallography.
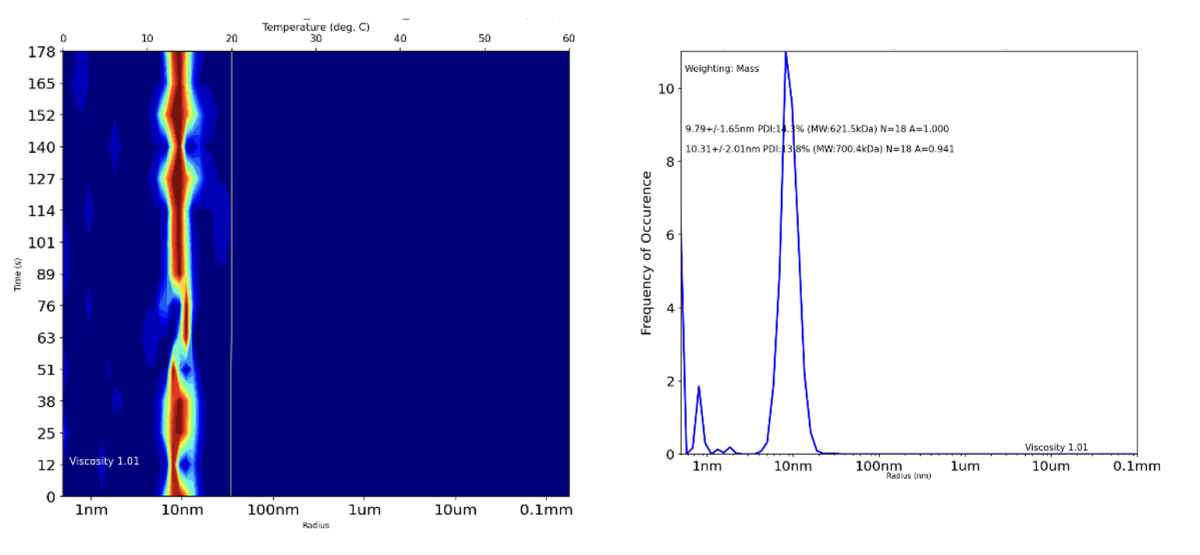
Figure 2: DLS data output for a PoLiPa-solubilised ion channel after affinity and size exclusion chromatography (SEC) purification. The result indicates that there is very little free polymer and that all the protein is likely to be solubilized within the polymer. Again, there is no evidence of aggregation over time, and therefore the protein is suitable for downstream applications such as Cryo-EM.
Start your next project with Domainex
Contact one of our experts today