- About
-
Solutions
-
Services
- Biosciences
- Chemistry
- Integrated Drug Discovery
- Computer Aided Drug Design
- Hit Identification
- Target Classes and Modalities
- Therapeutic Areas
-
A-Z
- A
- B
- C
- D
- E
- F
- G
- H
- I
- K
- L
- M
- N
- O
- P
- R
- S
- T
- V
- X
-
Services
- Library
- News & Events
- Careers
Covalent Inhibitors
An established mechanism of action gaining new momentum
Covalent inhibitors are compounds which bind to a target protein by reaction of an electrophilic functional group (“warhead”) with a nucleophilic residue (typically cysteine, serine or threonine) on a target protein.
During the period between 2000-2020, significant effort went into the design of covalent drugs to treat oncology indications. There was a widely held concern that covalently modifying proteins could cause autoimmune reactions, such as idiosyncratic liver toxicology, which was why non-oncology indications were typically not pursued. Advances in the field over the last few years have led to the re-emergence of covalent inhibitor programmes for non-oncology indications1.
Covalent inhibitors have some potential advantages over non-covalent inhibitors such as increased therapeutic duration and reduced frequency of drug dosing. Additionally, they may be able to inhibit target proteins once deemed undruggable (i.e. proteins with shallow binding pockets).
Covalent Drug Discovery at Domainex
At Domainex, our scientists are highly skilled and experienced in the design, synthesis and testing of both reversible and irreversible covalent inhibitors. We have an in-house covalent fragment library available and ready to screen to get your programme started. The library includes a diverse set of compounds (including both irreversible and reversible covalent warheads), pre-filtered to only include fragments with preferred covalent warheads (i.e. those found in approved drugs, investigational drugs in the clinic or gaining traction in the literature) and fragment-like properties. Each fragment has been manually inspected to ensure elaboration is synthetically tractable. Alternatively, Domainex can screen commercially available compound libraries or libraries provided by our clients.
At Domainex, covalent fragments are initially screened using a mass-spectrometry (MS) based assay. The binding percentage (ratio of bound protein to total protein) and the stoichiometry of binding observed for each fragment is reported.
For irreversible inhibitors, initial hits are then profiled in our glutathione (GSH) reactivity assay, which provides real time kinetics on the fragment's reactivity with freely available cysteine residues. Compounds with a half-life of <100 minutes are rejected as containing unfavourable highly reactive warheads which may react with off-target proteins and cause unwanted side effects. In addition to assessing irreversible inhibitors, our GSH reactivity assay has been adapted and optimised to identify the presence of reversible covalent inhibitors.
Selected covalent hits are progressed for binding site identification by liquid chromatography (LC)-MS peptide mapping to confirm covalent binding to the target amino acid. This technique is suitable for all irreversible inhibitors and reversible inhibitors which have a slow off rate. The binding site(s) are identified by utilising a bottom-up proteomics approach, where the fragment-protein conjugate is digested into peptides by a proteolytic enzyme (typically trypsin) and analysed by liquid chromatography-high resolution accurate mass spectrometry (LC-HRAMS).
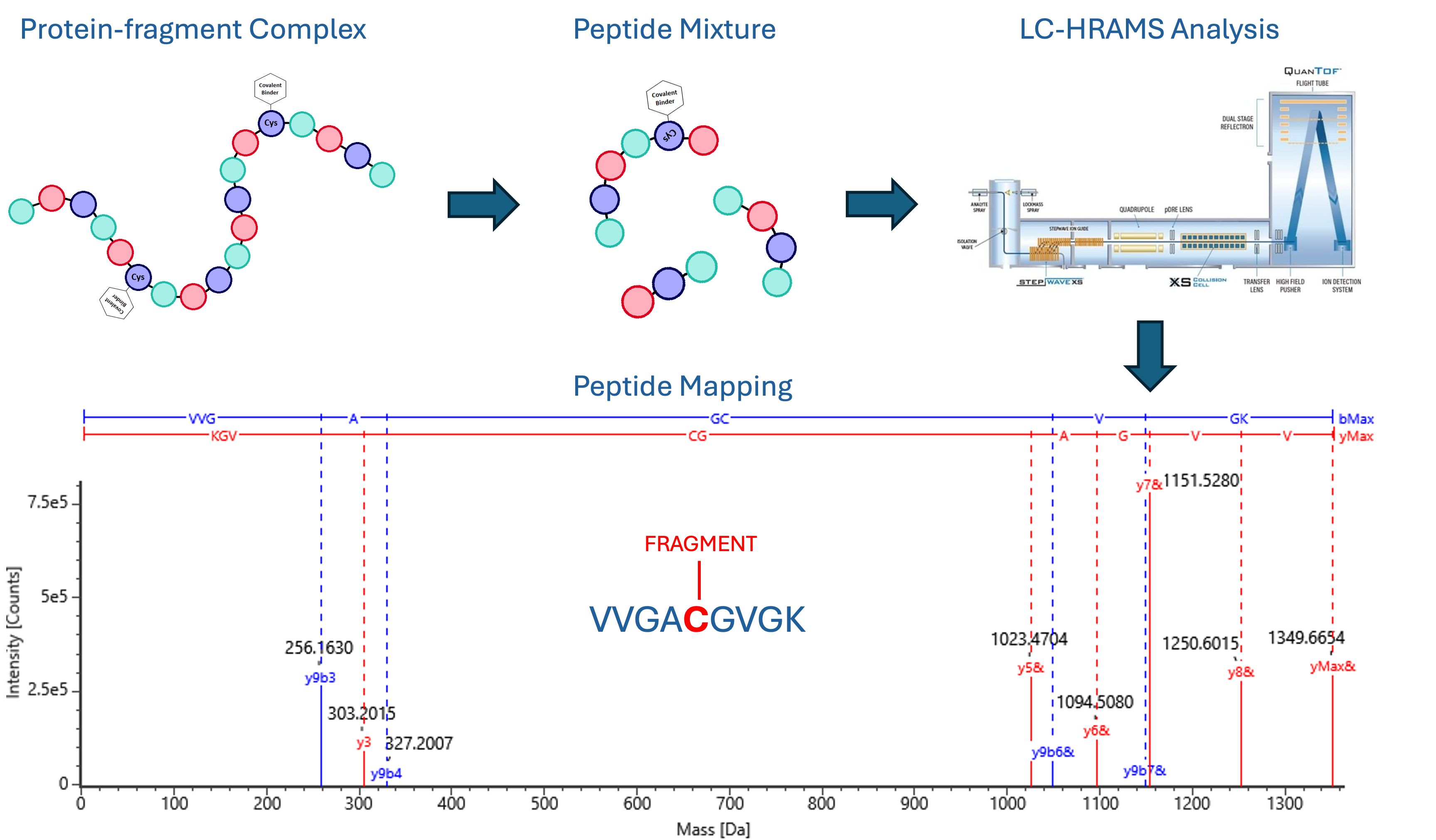
Figure 2: Identification of a covalent fragment binding to Cys12 of KRAS G12C protein using Domainex’s peptide mapping workflow
Additional fragment characterisation using our MS-based assay can be performed. For irreversible covalent inhibitors, the second order rate constant kinact/KI, which measures the potency of covalent binding can be determined2. kinact/KI is considered superior to an IC50 value for covalent inhibitors since it captures the inactivation in a time-dependent manner. For reversible covalent binders, the Kobs, the maximum percentage of binding, and Koff rate value can be determined and reported.
Once suitable chemical starting points have been identified for your project, we can provide an integrated team to perform hit-to-lead and lead optimisation campaigns. Our GSH reactivity assay, binding site identification methods and kinact/KI assay can continue to be deployed as your programme progresses, along with structural biology techniques to leverage a structure-based drug discovery (SBDD) approach. Additionally, our direct-to-biology (D2B) platform can be applied to rapidly generate structure activity relationships (SAR) by synthesising hundreds or thousands of compounds in plate-based format.
- S. E. Dalton; O. D. Pietro; E. Hennessy. A Medicinal Chemistry Perspective on FDA-Approved Small Molecule Drugs with a Covalent Mechanism of Action. J. Med. Chem. 2025, 68, 2307-2313
- K. S. Li; J. G. Quinn; M. J. Saabye; J. F. S. Guerrero; J. Nonomiya; Q. Lian; W. Phung; Y. Izrayelit; B. T. Walters; A. Gustafson; N. F. Enders; M. H. Beresini; M. M. Mulvihill. High-Throughput Kinetic Characterisation of Irreversible Covalent Inhibitors of KRASG12C by Intact Protein MS and Targeted MRM. Anal. Chem. 2022, 94, 1230-1239

Figure 1: Domainex’s irreversible covalent fragment screening workflow
Start your next project with Domainex
Contact one of our experts today