- About
-
Solutions
-
Services
- Biosciences
- Chemistry
- Integrated Drug Discovery
- Computer Aided Drug Design
- Hit Identification
- Target Classes and Modalities
- Therapeutic Areas
-
A-Z
- A
- B
- C
- D
- E
- F
- G
- H
- I
- K
- L
- M
- N
- O
- P
- R
- S
- T
- V
- X
-
Services
- Library
- News & Events
- Careers
Cryo-EM analysis of agonist-activated ion channel TrpML3
Generation of both apo and ligand bound Cryo-EM maps for TrpML3
Challenge
Ion channels are important drug discovery targets, involved in numerous diseases. However, as integral membrane proteins, they are a challenging class of proteins to investigate, especially by biophysical and structural approaches.
Our target of interest, TrpML3 (Mucolipin-3) (UniProt: Q8TDD5) is an ion channel implicated in autophagy, an intracellular degradation pathway which is induced by cellular stresses. Early clinical trials have demonstrated the feasibility and potential benefit of inhibiting autophagy in multiple cancer types (Levy et al.1). Inhibition of TrpML3 suppresses autophagy, while activation has the opposite effect (Kim et al.2), highlighting the potential benefit of targeting this protein in drug discovery programmes. Therefore, we wanted to generate a cryo-EM structure to enable structure-based drug discovery.
Solution
To study TrpML3, full-length protein was expressed in Expi293™ mammalian cells and purified in the detergent lauryl maltose neopentyl glycol (LMNG). Dynamic Light Scattering (DLS) analysis confirmed a single monodisperse population had been obtained (Figure 1).
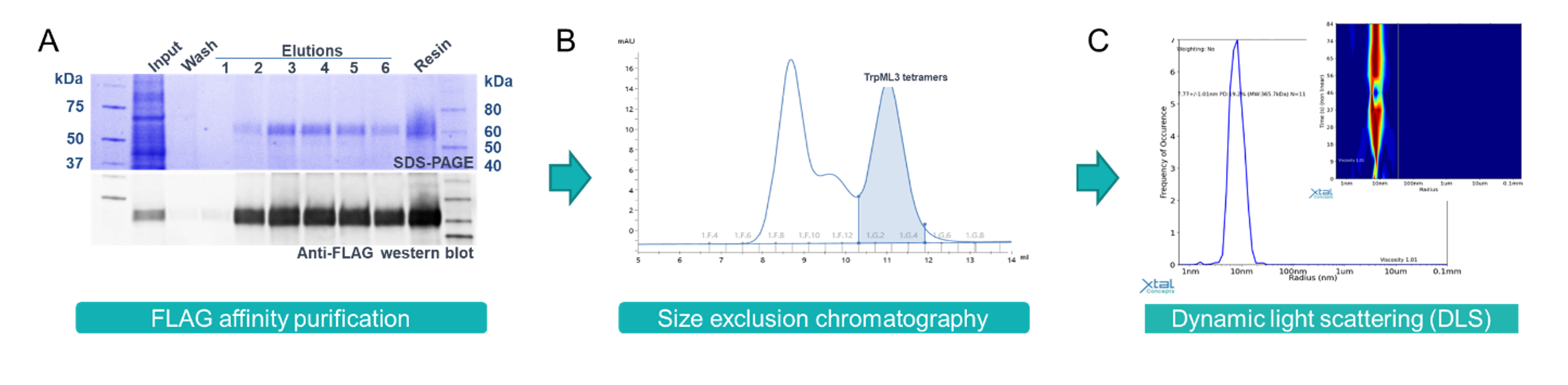
Figure 1: Purification of LMNG detergent solubilised TrpML3. A) SDS-PAGE and anti-flag western blot showing affinity purified TrpML3. B) Size exclusion chromatography of TrpML3 with a peak present at the expected size for the tetrameric ion channel. C) DLS analysis confirming a monodisperse population of TrpML3 protein ready for Cryo-EM studies.
TrpML3 cryo-EM grids were prepared in the presence and absence of ML-SA1 (a known agonist of TrpML3). First, freezing conditions were screened to identify grids with consistent protein distribution over a sufficient area of the grid. The most promising grids were progressed for data collection on a 300 KeV Titan Krios microscope with K3 detector. In a single 24-hour session, a total of ~5,000 micrographs were collected for the apo TrpML3 grid, while ~3,500 micrographs were collected for TrpML3 in the presence of ML-SA1 for data processing.
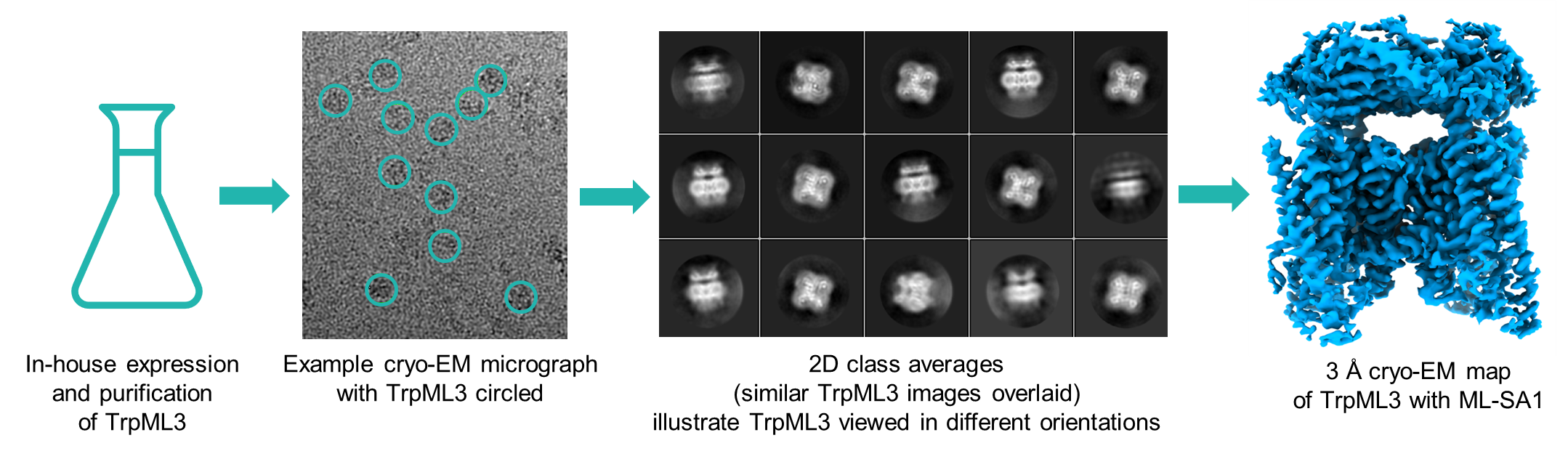
Figure 2: Flowchart for Cryo-EM analysis of TrpML3 2D class averages showed a mixture of different orientations for the tetramer (Figure 2), with secondary structure visible, confirming the presence of high-resolution information within the dataset. 3D refinement of the full complex showed that the apo TrpML3 was in a closed and inactive conformation, whereas in the presence of agonist ML-SA1, the channel was open. The resultant re-arrangement in helices between the open and the closed conformation could be observed even at low resolution (Figure 3 top). Moreover, unlike the previously published structures (Zhou et al.3), the resultant high-resolution maps enabled identification of the bound ligand, as well as protein side chain conformations. (Figure 3 bottom).
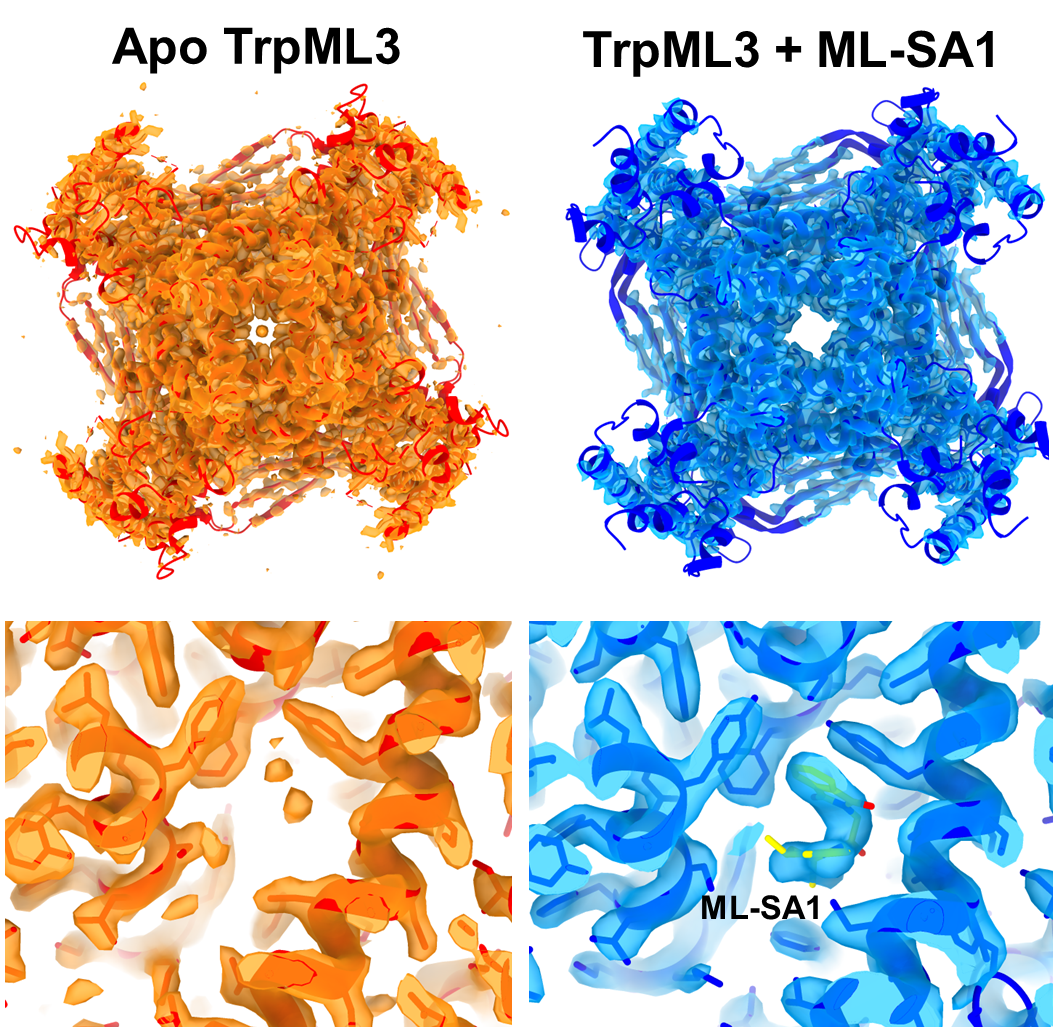
Figure 3: Comparison of post-processed electron-potential maps of apo (orange, 2.7 Å) and ML-SA1 bound TrpML3 (blue, 3.0 Å). Top, A transition from the closed TrpML3 channel apo state to the open channel ML-SA1 bound state is observed. Bottom, Comparison of the ligand binding pocket of apo-TrpML3 with ML-SA1 bound TRPML3 highlights the inclusion of ML-SA1 (yellow sticks), with side chain interactions leading to TRPML3 activation. The apo and ligand-bound TrpML3 structures have enabled computer-aided drug design (CADD) and virtual screening.
Figure 4: TrpML3 visualization. The video highlights structural differences between apo TrpML3 (orange) in a closed, inactive state; and ML-SA1 bound TrpML3 (blue), in an open conformation. Focusing on the ML-SA1 binding site of the open TrpML3 map, density is present for ML-SA1 (shown as a stick model), which is absent in the apo map.
Conclusion
Following successful membrane protein purification, structural biologists at Domainex generated both apo and ligand bound Cryo-EM maps for TrpML3, at 2.7 Å and 3.0 Å resolution, respectively. Comparison of the two derived atomic models demonstrated a clear transition from the closed channel apo state to the open channel ligand bound state (Figures 3 and 4). Detailed map features in the region of the ligand and its binding pocket have been used as a guide to model ligand location and conformation (Figure 4), thereby opening the way for future CADD and virtual screening efforts.
References
Levy, J. M. M., Towers, C. G., & Thorburn, A. (2017). Targeting autophagy in cancer. Nature Reviews Cancer, 17(9), 528-542
Kim, H. J., Soyombo, A. A., Tjon‐Kon‐Sang, S., So, I., & Muallem, S. (2009). The Ca2+ channel TRPML3 regulates membrane trafficking and autophagy. Traffic, 10(8), 1157-1167
Zhou, X., Li, M., Su, D., Jia, Q., Li, H., Li, X., & Yang, J. (2017). Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states. Nature structural & molecular biology, 24(12), 1146-1154.
Start your next project with Domainex
Contact one of our experts today