- About
-
Solutions
-
Services
- Biosciences
- Chemistry
- Integrated Drug Discovery
- Computer Aided Drug Design
- Hit Identification
- Target Classes and Modalities
- Therapeutic Areas
-
A-Z
- A
- B
- C
- D
- E
- F
- G
- H
- I
- K
- L
- M
- N
- O
- P
- R
- S
- T
- V
- X
-
Services
- Library
- News & Events
- Careers
Adenosine A2a receptor: Novel Biophysical Fragment Screening using Polymer-Encapsulated Nanodiscs
Introduction
Fragment screening is a useful method for Hit Identification as it offers an efficient and flexible approach to identify novel therapeutic starting points that other techniques may have missed due to the small molecular size of the fragments. However, fragment screening usually requires access to purified proteins which can be difficult to obtain when working with membrane proteins such as G protein-coupled receptors (GPCRs).
Domainex’s PoLiPa (Polymer Lipid Particle) technology stabilises membrane proteins by encapsulating the target protein in a polymer that encloses a small disc of the native cell membrane lipids, known as a nanodisc.
For the first time, Domainex has demonstrated that polymer nanodics can be used in conjunction with Spectral Shift for fragment screening – expanding the targets that fragment screening can be used for and opening up the potential to develop novel therapeutics agents.
Challenge
GPCRs are important drug discovery targets and are a challenging class of proteins to produce and purify in multi-milligram quantities. The poor yields associated with GPCRs are typically caused by low levels of expression, poor extraction efficiency and stability issues. Traditionally, detergents are used to extract receptors by forming detergent micelles to stabilise them. However, these micelles are not representative of the membrane protein’s natural environment, often display low levels of activity and can interfere with downstream applications, making fragment screening against GPCRs a particular challenge.
Solution
At Domainex, our researchers have optimised an alternative, detergent-free strategy to purify membrane proteins, such as GPCRs, ion channels and solute carriers. Synthetic polymers are available that can solubilise cell membranes and spontaneously form nanodiscs containing both native lipids and membrane proteins. These polymer-encapsulated nanodiscs can be formed with several different amphipathic co-polymers, composed of alternating hydrophobic and hydrophilic groups, such as styrene maleic acid (SMA), diisobutylene-maleic acid (DIBMA), styrene-acetic acid (AASTY), cycloalkane modified amphipols and others. Collectively, we refer to these polymer-encapsulated nanodiscs as PoLiPa. A key advantage of this extraction method is the ability to maintain the natural lipid composition around the membrane protein, which is representative of the native environment and may be important for physiological folding and functionality of the protein.
Here, we show that polymer-encapsulated nanodiscs of A2a are physiologically folded and suitable for biophysical ligand binding applications.
The class A GPCR, Adenosine A2a receptor (UniProt ID P29274), was expressed in insect cells using a baculoviral expression system. The GPCR was then purified using a PoLiPa polymer formulation rather than using traditional detergent extraction methods. The binding of three known small molecule antagonists was confirmed by nano differential scanning fluorimetry (nanoDSF) showing that the polymer-purified Adenosine A2a receptor was stable and physiologically folded. (Figure 1).
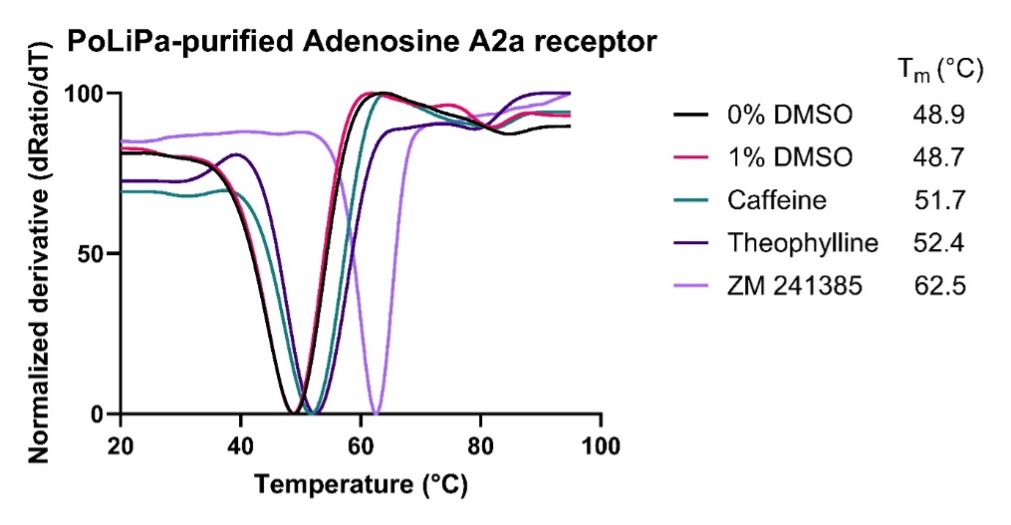
Figure 1: Thermal stability of PoLiPa purified Adenosine A2a receptor in the presence of DMSO and three known small molecule antagonists (caffeine, theophylline and ZM241385).
Polymer-encapsulated nanodiscs are relatively large and, depending on the polymer used, can vary between 9 to 25 nm wide. The size of the protein containing nanodisc can limit sensitivity in methods where immobilisation of the nanodisc is required for ligand binding studies such as surface plasmon resonance (SPR) or grating coupled interferometry (GCI). Spectral Shift technology from NanoTemper, measures direct ligand binding to the target in solution and monitors ligand binding through changes in fluorescence. These changes in the emission profile are typically induced by ligand binding altering the chemical environment of the dye, either through proximity or conformational changes. Spectral Shift does not typically suffer from the same limitations of size or buffer composition as other techniques and multiple labelling strategies are compatible with Spectral Shift. Using this technique, it is possible to measure shifts in the emission profile of a fluorescent tag bound to the Adenosine A2a protein.
The Adenosine A2a receptor was expressed with a His-tag and successfully labelled using the non-covalent affinity-based NanoTemper RED-tris-NTA dye. A Spectral Shift assay was developed and validated by measuring the affinity of four known A2a receptor antagonists. The affinities determined were in accordance with literature values (Figure 2).

Figure 2: Affinity determination of four known antagonists (theophylline, caffeine, ZM241385 and DPCPX) of Adenosine A2a receptor using Spectral Shift.
Following assay validation, a pilot screen was performed using a subset of Domainex’s in-house fragment library; 160 fragments were tested at a single concentration (250 µM) with theophylline used as a positive control. The pilot screen was successful with a hit rate of 9.38%, using a cut-off based on the positive control (≥70% Δratio of theophylline binding) (Figure 3).
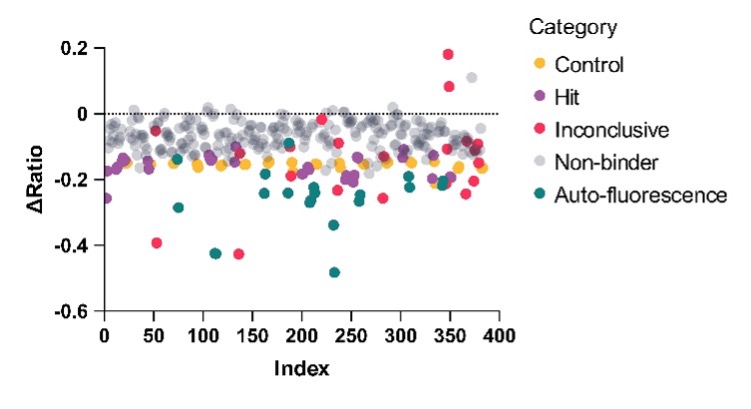
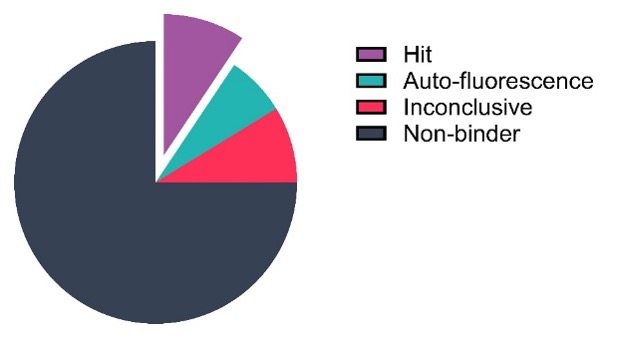
Figure 3: Single concentration screen of 160 fragments against the Adenosine A2a receptor using Spectral Shift. Data visualised in scatter plot and pie chart formats.
Following the successful pilot screen, a further 800 fragments were screened, using the same method, resulting in a total of 125 initial hits being identified. A second single-concentration screening step at 100 µM was performed, to eliminate the weaker hits and select the best binders for affinity determination. A total of 31 fragments were selected and screened using a 12-point 1:1 dilution series from a top concentration of 250 µM (Figure 4A). The pKD and ligand efficiency values were determined for the final set of hits (Figure 4B), identifying 19 promising hits with ligand efficiencies over 0.3.
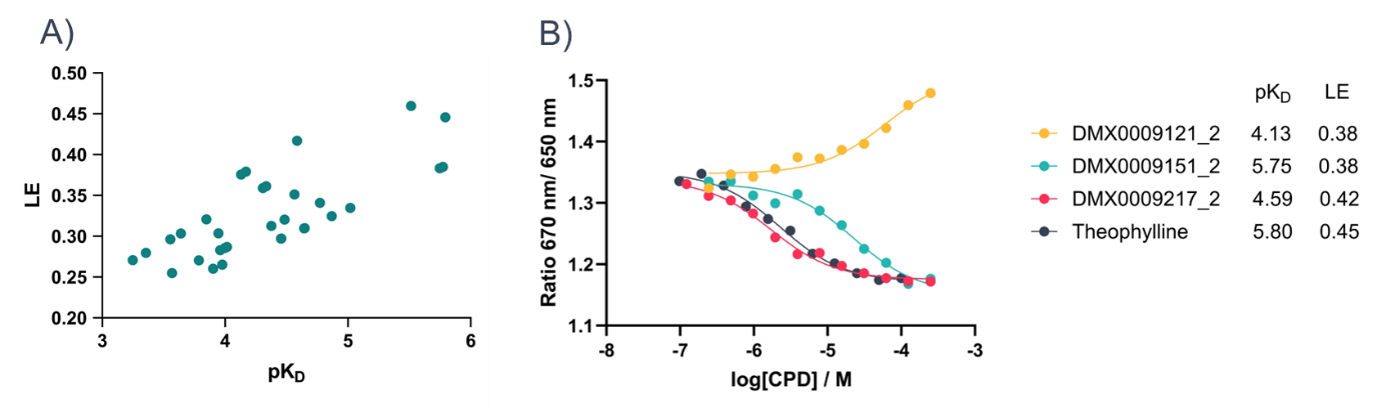
Figure 4: A) pKD vs ligand efficiency scatter plot of fragment hits. B) Example concentration response curves of fragment hits in comparison to the known antagonist theophylline.
Conclusions
The Adenosine A2a receptor was successfully purified using polymer-encapsulated nanodiscs. Physiological folding and stability of the membrane protein were validated using nanoDSF and Spectral Shift. A total of 960 fragments were screened, leading to the identification of several promising hits for further study and characterisation, highlighting the compatibility of PoLiPa and Spectral Shift and the power of combining the two techniques. We believe this is the first time polymer-encapsulated native nanodiscs have been used for fragment screening; a significant advance in hit identification methods for membrane protein drug discovery research.
Start your next project with Domainex
Contact one of our experts today