- About
-
Services
-
Offerings
- Offerings
- ADME and Bioanalytical Sciences
- Analytical Chemistry
- Assay Development
- Biochemical Assays
- Biophysical Assays
- Cell Based Assays
- Computational Chemistry
- Fragment and Compound Screening
- Integrated Drug Discovery Services
- Medicinal Chemistry
- Project Management and Consultancy Services
- Protein Expression and Purification Services
- Structural Biology
- Synthetic Chemistry
- Virtual screening
-
Research Phases
- Research Phases
- Hit Identification
- Hit to Lead
- Lead Optimisation
- Therapeutic Areas
- Target Classes
-
Approaches & Techniques
- Approaches & Techniques
- CDH (Target Gene Fragmentation)
- Cryogenic Electron Microscopy (Cryo-EM)
- Differential Scanning Fluorimetry (DSF) and nanoDSF Services
- Direct-to-Biology (D2B)
- Dynamic Light Scattering (DLS)
- eProtein Discovery
- Flow Cytometry
- Fragment Based Drug Discovery (FBDD)
- FragmentBuilder
- Grating-Coupled Interferometry
- High Throughput Screening
- Isothermal Titration Calorimetry (ITC)
- LeadBuilder
- PoLiPa (Membrane Protein Solubilisation)
- Spectral Shift and MST Services
- Structure Based Drug Design (SBDD)
- Surface Plasmon Resonance (SPR)
- X-ray Crystallography
-
Offerings
- Library
- News & Events
- Careers
Targeted Protein Degrader Toolbox
Accelerate your research with Domainex's toolbox
To expedite targeted protein degrader programmes, Domainex has generated a PROTAC® toolbox consisting of approximately 160 partial PROTAC compounds (comprised of a linker and an E3 ligase binder). The toolbox is designed to be combined with Domainex’s Direct-to-Biology (D2B) platform for the rapid, plate-based synthesis of PROTACs. Each partial PROTAC compound is comprised of an E3 Ligase recruiter ligand and a linker which includes a synthetic handle. The majority of the compounds are not commercially available from other vendors and are ready for immediate reaction with a protein of interest (POI) binder.
PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license
E3 Ligase Recruiters
To date most R&D in the protein degrader field has focussed on exploiting Cereblon (CRBN) and Von Hippel-Lindau tumour suppressor (VHL) E3 ligases. Where structural information is available, over half of the current clinical stage PROTACs utilise CRBN based ligase recruiters, where the beyond rule-of-5 (bRo5) molecular property space is more favourable for optimisation and delivery of oral bioavailable degraders.1
However, for CRBN ligase recruiters, selectivity for degradation of the protein of interest (POI) versus proteasomal degradation of neo-substrates, where the CRBN ligase recruiter acts as a molecular glue, remains a challenge. Several neo-substrates are known to play important physiological roles and their protein degradation can often lead to toxicological consequences. As an example, degradation of Spalt Like Transcription Factor 4 (SALL4) by thalidomide resulted in teratogenicity and the epidemic of severe birth defects in the late 50’s-early 60’s.
Given the interest in CRBN from a clinical perspective, our toolbox is biased towards CRBN-based recruiters, but with a significant subset designed to reduce potential neo-substrate protein degradation (Figure 1).
Linkers
It is well documented that both the linker length and its composition play an instrumental role in driving protein degradation of the POI and contributing to the property profile. As a consequence, Domainex’s toolbox includes partial PROTACs with a range of linker types including variation in length, rigidity and molecular property space (rotatable bonds, PSA, clogP, MW, ionisation state) (Figure 2). Different exit vectors (composition, position…) and multiple synthetic handles ready for coupling with POI ligands bearing different functionality are also included.
Reference
- Yang et al., Drug Discovery Today, Volume 29, Issue 2, February 2024, 103865.
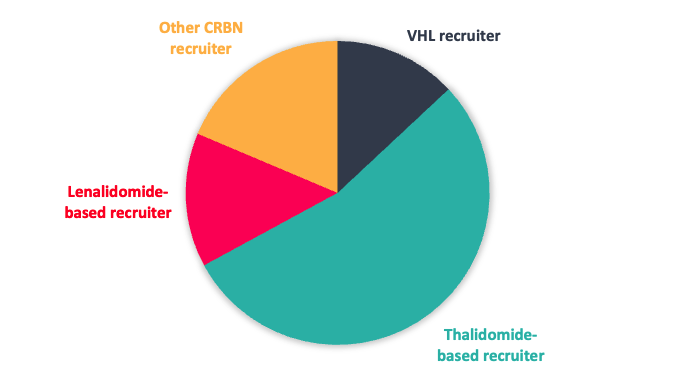
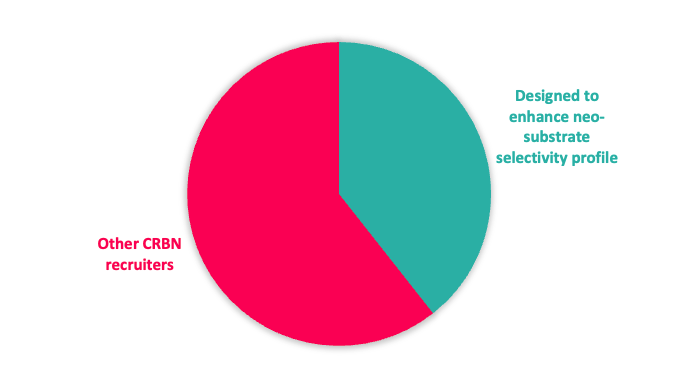
Figure 1: Top; E3 ligase recruiter composition. Bottom; proportion of CRBN recruiters designed to reduce neo-substrate protein degradation
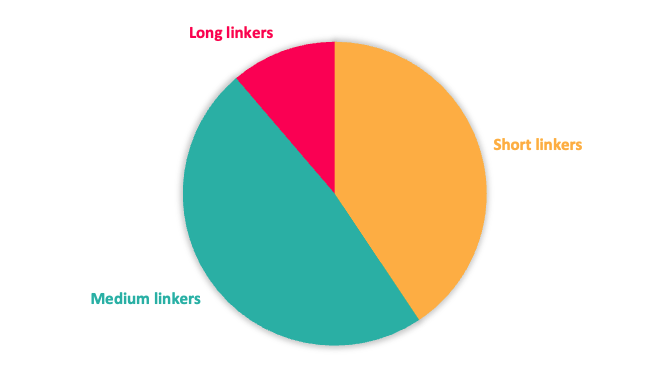
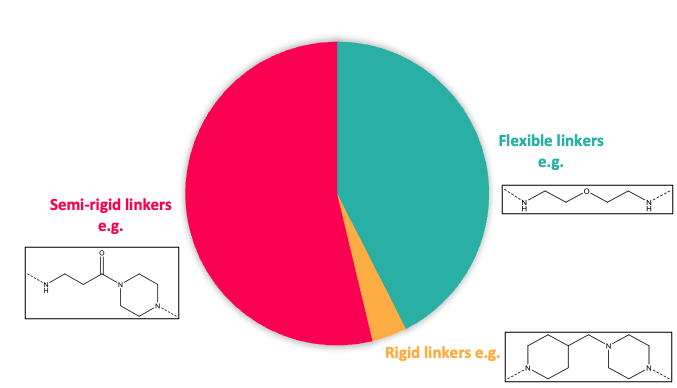
Figure 2: Top; Toolbox linker length composition. Bottom; Toolbox linker rigidity
Case Study
Target: Aurora Kinase (overexpressed in human tumors)
JB170 is a potent and highly specific CRBN mediated Aurora A degrader compound using Alisertib as the POI ligand. As a pilot study, the acid of Alisertib was coupled to 40 amine representative examples from Domainex’s toolbox.
The plate-based chemistry was completed in less than 3 days including automated LC-MS analysis, which showed > 75% of the requisite amides were synthesized in acceptable purity. Profiling of the unpurified mixtures in a HiBiT tagged Aurora A degradation assay revealed several known and novel linker based CRBN derived compounds with DC50 values in the 30-150 nM range, similar to that observed with JB170 (DC50 28 nM) (Table 1).
Summary
Selected partial PROTACs from our toolbox were successfully used in plate-based chemistry reactions and progressed directly to a HiBiT degradation assay. Several hits, with both known and novel linkers, were identified, demonstrating the benefit of the library when combined with a D2B approach.


Table 1: Selected hits identified from the HiBiT screen
Start your next project with Domainex
Contact one of our experts today