Written by Alicia Overall (Scientist) and Dillon J. Rinauro (Senior Scientist I), with support from Alec O’Keeffe (Associate Principal Scientist) and Kartika Shetty (Senior Scientist II), who all work within Domainex’s biophysical assay team.
Drug discovery is an inherently expensive and time-consuming process. A fully realised therapeutic can cost hundreds of millions of dollars and take 10-15 years to appear on the market, if it gets through clinical trials successfully at all. (1,2)
In the early stages of discovery, drug developers need to demonstrate and understand the engagement of their compound of interest with its target. Failure to establish evidence of contact between the two binding partners can be a major setback for lead characterisation. A lead candidate may demonstrate a promising capacity to achieve a desired effect, but if no evidence exists for direct target engagement, then more resources will be needed to resolve the mechanism of action, introducing further time and money constraints. It is to the pharmaceutical industry’s advantage to take a biophysical approach to early-stage drug discovery to mitigate these issues.
Biophysics can be used to answer questions such as: How quickly does a drug candidate bind to its target? Where does the candidate bind? How long does this interaction last? Is the interaction enthalpically or entropically driven? As biophysical scientists, here are five biophysical assays that we routinely deploy to build a more comprehensive understanding of drug-target interactions.
Differential scanning fluorimetry (DSF) is a medium-throughput, solution-based technique that measures the relative folded state of a protein. Heat energy is supplied to the protein, increasing the range of motion until it ‘breaks free’ from its native conformational state. This unfolding process is monitored by either the intrinsic fluorescence of amino acids or the use of a fluorescent dye. The melting temperature (Tm), the point at which 50% of the protein is unfolded, is likely to increase if the drug candidate molecule successfully binds to the target as this stabilises the conformation.
Unfortunately, DSF can only really inform a user of target engagement; it is limited in its capacity to provide more detailed binding information, and therefore should ideally be supplemented by one of the methods described below.
Microscale thermophoresis (MST) is higher throughput than most other biophysical methods and utilises the response of a fluorescent molecule or protein when a temperature gradient is applied. The response change depends on the concentration of a binding partner and can be quantified using TRIC (temperature related intensity change) or thermophoresis (rate of motion across the temperature gradient). In general, the fluorescently labelled molecule will move across the temperature gradient and the rate at which it does so depends on its size, while the intensity is influenced by the changes in the fluorescent dye microenvironment. Hence, changes in the intensity and rate of movement can inform us of the relative bound/unbound state of two binding partners, and therefore the affinity (KD)between these two binding partners.

Some MST instruments are now also equipped with Spectral Shift technology. This technique, now available at Domainex, employs precision optics to measure wavelength shifts induced due to the binding of a fluorescently labelled biomolecule with a ligand. The principles are similar to MST but with improved sensitivity as the measurements are ratiometric and hence free from pipetting biases that are induced in absolute fluorescence intensity measurements.
To gain further insights into the binding interaction, isothermal titration calorimetry (ITC) can be employed. Though lower throughput compared to other methods, it offers an advantage because in addition to affinity, it also reveals binding stoichiometries and the thermodynamics of these binding interactions. The technique measures the microscopic amount of heat released (or absorbed) upon the binding of a potential therapeutic to a biomolecule to tell us whether the binding event is enthalpically driven. The main limitations of ITC are throughput and sample consumption; however, ITC provides invaluable information during the drug discovery process, especially when binding thermodynamics are combined with ligand-bound protein structures

One more recently developed method that pairs well with ITC is grating-coupled interferometry (GCI). GCI uses a surface-based, label free technique to measure the binding kinetics of a drug candidate, the analyte, and an immobilised/captured target, the ligand. With clog-free microfluidics, the analyte flows over the surface where the ligand is bound. If binding occurs, the relative mass on the surface will increase, acting like a microscopic scale. With knowledge of the molecular weights, GCI goes above and beyond methods like MST and DSF by not only determining the affinity of two binding partners, but also the association (kon or ka) and dissociation (koff or kd) rate constants.
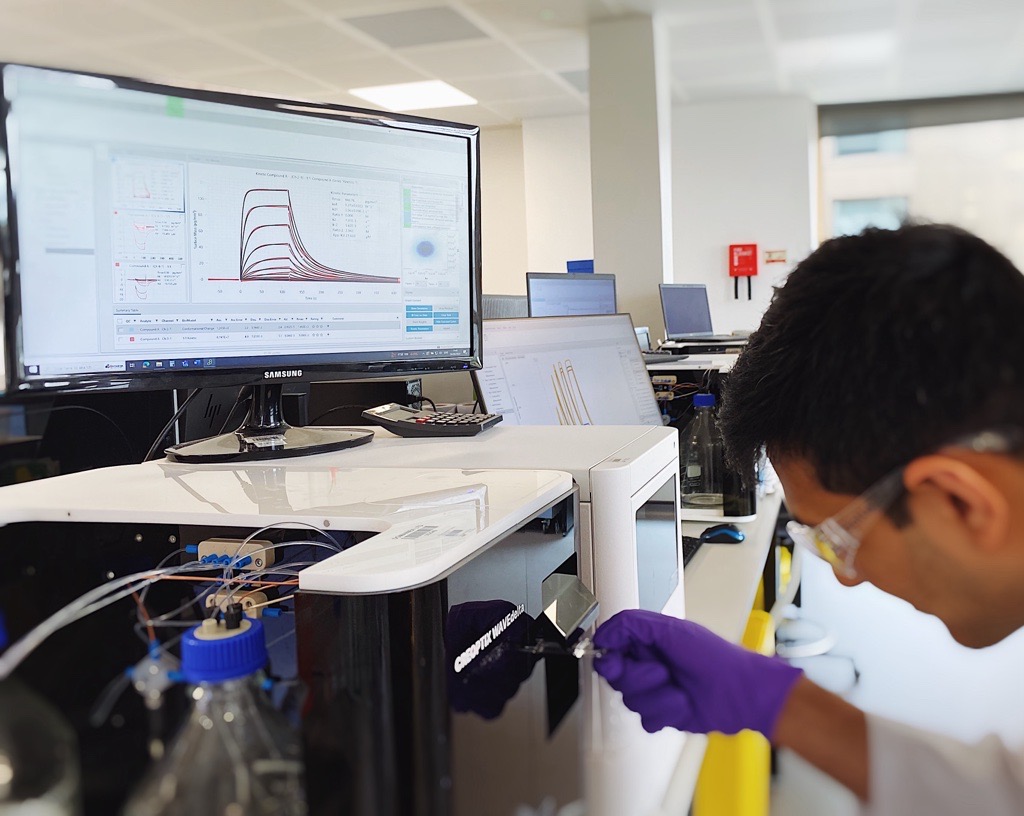
Perhaps the strongest advantage of GCI is its RAPID™ (Repeated Analyte Pulses of Increasing Duration) kinetics mode. Rather than measuring several different concentrations of the analyte for a set period of time, GCI instead holds constant the concentration of analyte whilst varying the time of interaction. In practice, GCI has a much higher potential for increased throughput when compared to basic Surface Plasmon Resonance (SPR) systems which are also generally more expensive. Further advantages of GCI include: the WAVEguide system, which samples across the entire flow cell and therefore increases instrument sensitivity, clog-free microfluidics, and the ability to measure faster association and dissociation rates to measure the binding kinetics of weaker compounds such as fragments.
With its RAPID™ kinetics and waveguide technology for enhanced sensitivity, GCI is well adapted for implementation in the screening of low-molecular weight molecules, particularly within fragment-based drug discovery (FBDD) programmes. FBBD can drastically improve the success rate of a drug pipeline compared to traditional high throughput screening (HTS).
A major pharma company recently reported only five leads from over 70 campaigns with traditional HTS methods. On the other hand, FBDD has successfully generated potent inhibitors of targets previously thought to be undruggable, even though three to four orders of magnitude fewer compounds are screened compared to HTS. Kirsten Rat Sarcoma Viral Oncogene Homologue (KRAS), an important target in cancer research, has been deemed a challenging target; that is, until recent FBDD campaigns generated promising results in clinical trials.(3)
So why has FBDD proven to be so promising for drugging notoriously difficult targets? Generally, fragments bind to proteins with weak affinities (micromolar range); however, their smaller nature compared to molecules from HTS campaigns typically have much higher intrinsic binding energies and take advantage of potential binding space within a pocket. Biophysical methods like GCI and ITC offer valuable insight into affinity, kinetics and thermodynamic parameters in early biophysical characterisation of compounds. This additional information enables us to build a detailed picture of a molecule’s characteristics and specific interactions to serve as a benchmark throughout the optimisation process and help gauge whether efforts have successfully improved the potency of the lead compound.
The world of drug discovery is an ever-changing landscape, and biophysics is one of many fields pushing the boundaries of what is possible. Domainex has seen significant expansion and development in its biophysics team over the last decade and these techniques are now providing answers to questions, sometimes before they have even arisen - truly highlighting the importance that biophysics holds within drug discovery. Utilising these varied techniques helps to drive drug discovery, especially in conjunction with FBDD, to open up once ‘undruggable’ targets. These trends will continue to help shape the future of research.
Reference:
- Wouters, O. J., McKee, M., & Luyten, J. (2020). Estimated research and development investment needed to bring a new medicine to market, 2009-2018. Jama, 323(9), 844-853.
- Corr, P., & Williams, D. (2009). The pathway from idea to regulatory approval: examples for drug development. Conflict of interest in medical research education and practice.
- McAulay, K.; Bilsland, A.; Bon, M. Reactivity of Covalent Fragments and Their Role in Fragment Based Drug Discovery. Pharmaceuticals 2022, 15, 1366.