Advancing Compound Screening: An Introduction to Direct-to-Biology (D2B) Approaches
D2B represents a paradigm shift in compound screening methodologies. It offers a means to evaluate libraries of compounds directly using crude reaction mixtures, circumventing the time and resource-intensive purification steps that traditionally accompany library generation. This approach holds immense promise,1-3 significantly advancing the pace of compound screening efforts.
Requirements for Effective D2B Chemistry Methodologies
Critical to the success of D2B applications is the assurance of relatively clean reaction profiles. The accuracy of biological assay readouts relies upon compound mixtures containing minimal contaminants and unwanted by-products. This prerequisite underscores the need for optimised transformations which deliver target compounds in uncomplicated crude reaction mixtures which minimise contaminants that would be deleterious to the biological assay.
Recent Advances and Future Directions
A commonly used transformation for D2B screening is amide coupling, due to reliable product formation and relatively benign by-products. Indeed, Domainex has previously synthesised and screened a library of bifunctional degrader compounds (consisting of an E3 ligase recruiter, a linker and a protein of interest binder) in a D2B experiment (Figure 1a).
Recent literature has showcased significant strides in the field, with researchers demonstrating that Cu-catalysed azide-alkyne cycloaddition (CuAAC) platforms can be used to forge triazole linkages in a D2B approach (Figure 1b).4 The report not only demonstrated the compatibility of CuAAC mixtures with cellular assays, but also identified novel kinase binders using the technology. While extremely promising, the requirement for libraries of purified azide precursors poses a barrier in terms of accessibility and cost-effectiveness.
In light of these challenges, we hypothesised that an azidation reaction to convert amine libraries to the corresponding azides could be combined in situ with existing D2B-validated CuAAC conditions in a two-step one-pot azidation-CuAAC reaction (Figure 1c). This approach aims to enable the structural elaboration of cheaper and more readily available amine libraries to triazole linkages, offering an alternative approach to amide formation.
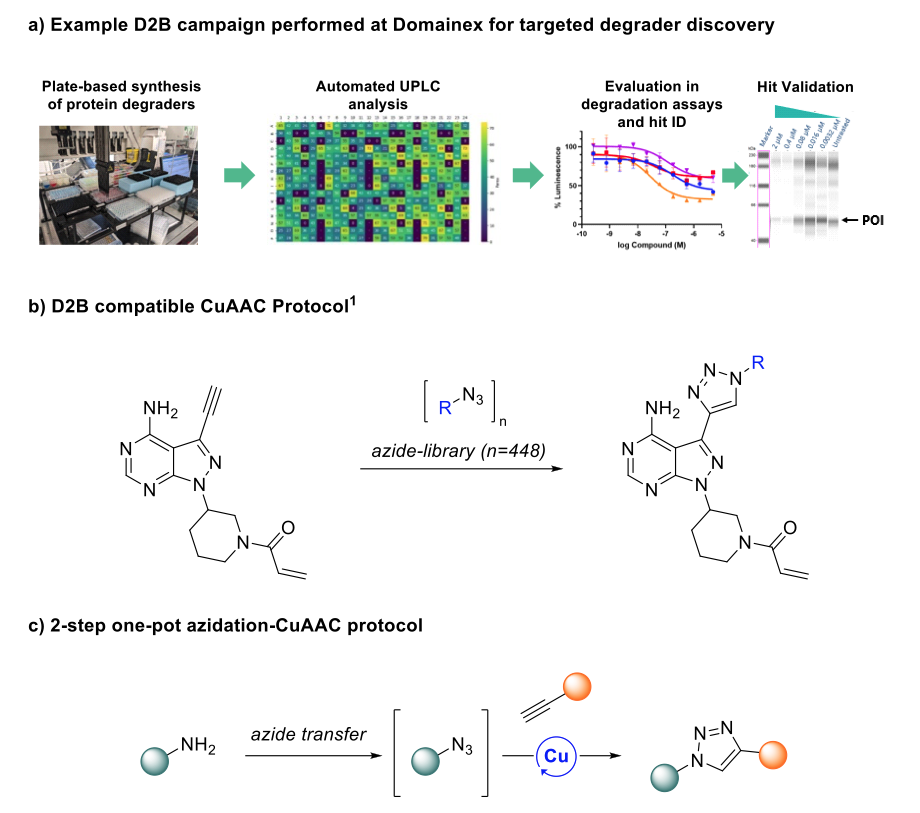
Figure 1: a) Example D2B campaign performed at Domainex. b) Reaction scheme showing CuAAC method recently validated for D2B library synthesis and cell-based screening, as reported by Gehrtz and co-workers.4 c) Novel 2-step one-pot azidation/CuAAC platform for D2B library screening, developed at Domainex.
Platform Development
In order to validate this methodology as a D2B compatible protocol, we carried out a multifactorial optimisation in a plate-based format to find the optimal combination of base, sodium ascorbate, and copper loading. This process allowed us to improve the yield of our model system from 3% to 55% (Figure 2).
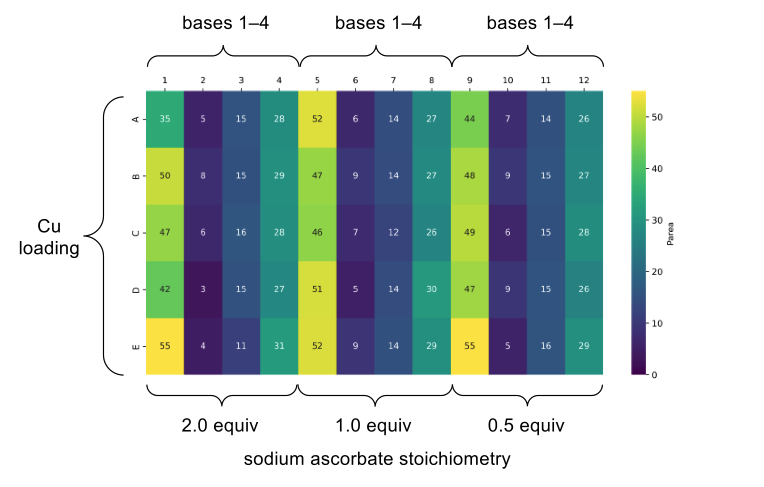
Figure 2: Plate-based optimisation of the two-step one-pot azidation/CuAAC reaction. This heatmap represents a multifactorial optimisation of base, copper loading and sodium ascorbate loading on a model reaction. Values correspond to UPLC product peak area.
To assess the robustness of the optimised protocol, a small library of 12 amine precursors, each bearing different functional groups, were combined with a model alkyne under the reaction conditions. Encouragingly, every amine was converted to the corresponding triazole in a clean reaction profile, with LCMS product peak areas up to 85% (Figure 3).
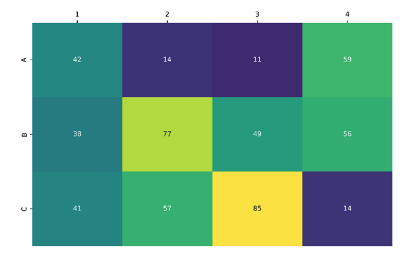
Figure 3: Heatmap demonstrating conversion of a panel of 12 amines to the corresponding triazole. Formation of the desired product was observed in every well.
This panel of crude triazole products was also submitted to a HepG2 cell viability assay alongside a control mixture which contained only the copper-click reagents but no substrate. As expected the copper control mixture displayed no cell toxicity. Furthermore, of the 12 crude reaction mixtures submitted, only 3 displayed measurable EC50 values (<50 μM) which was thought to be due to the triazole compounds synthesised (Figure 4). This result not only indicated that the reagents employed (and by-products formed) are not deleterious to biological assays, but demonstrated that this approach has the potential to identify hit compounds.
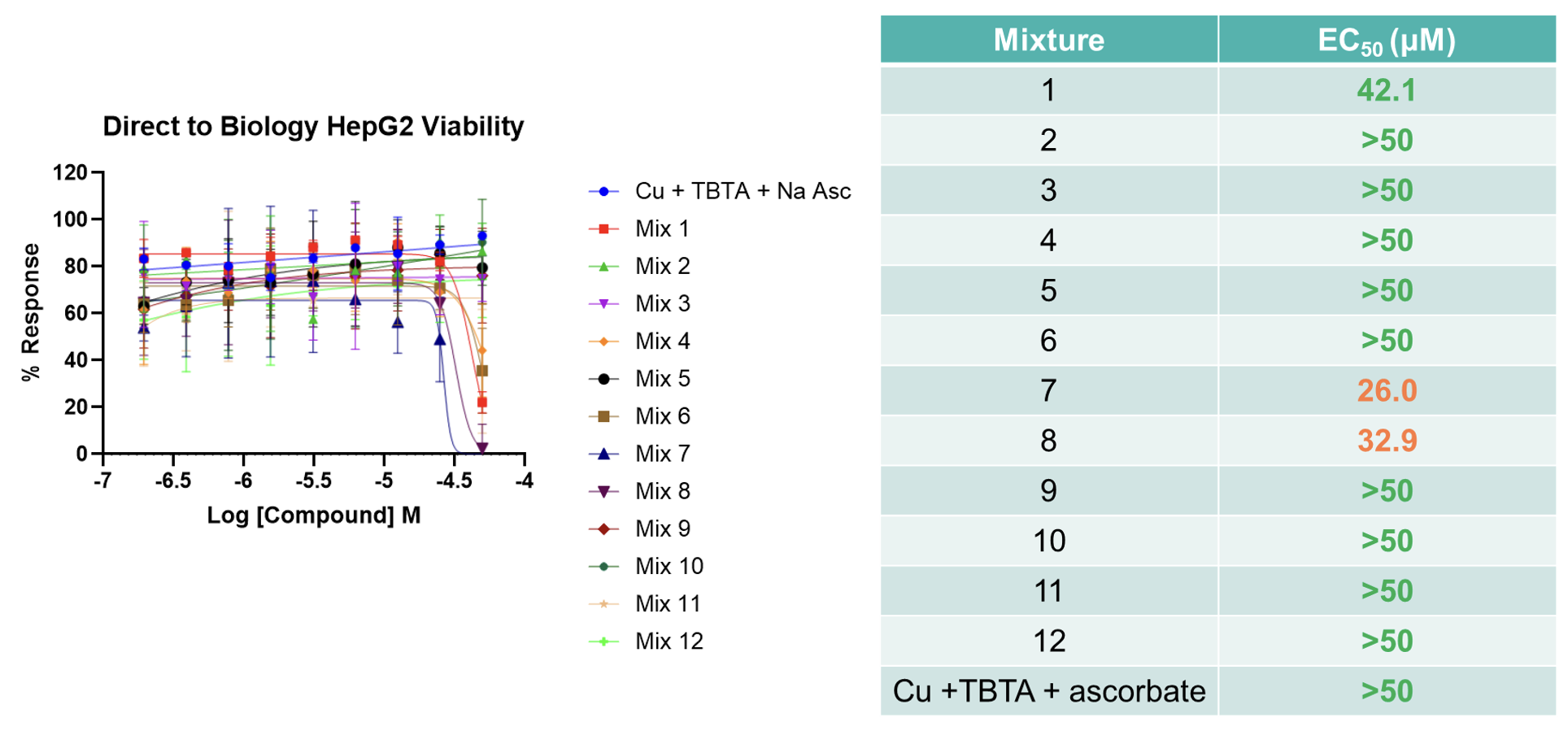
Figure 4: The 12 triazole products synthesised in our validation assay were assessed in a HepG2 cell viability assay (the EC50 values measured are provided in the table).
Application to Domainex’s Targeted Protein Degrader Toolbox
Having developed a robust platform for a two-step one-pot azidation/CuAAC with high tolerance for a range of chemical functionality and excellent compatibility with cellular assays, we next demonstrated that we could derivatise our proprietary targeted protein degrader toolbox with triazole linkers, in order to complement the previously established amide coupling protocol.
Domainex’s Targeted Protein Degrader toolbox
Approximately 160 partial degrader compounds (comprised of a linker and an E3 ligase binder)
Majority of library non-commercial
CRBN and VHL ligands
Variety of linkers and types
Different exit vectors
Ready for immediate coupling
We handpicked 36 partial degrader compounds from our in-house library and linked them to a model alkyne using our optimised protocol. This resulted in successful triazole functionalisation of all 36 partial compounds (100% success rate), with LCMS product peak areas ranging from 32–73% (Figure 5).
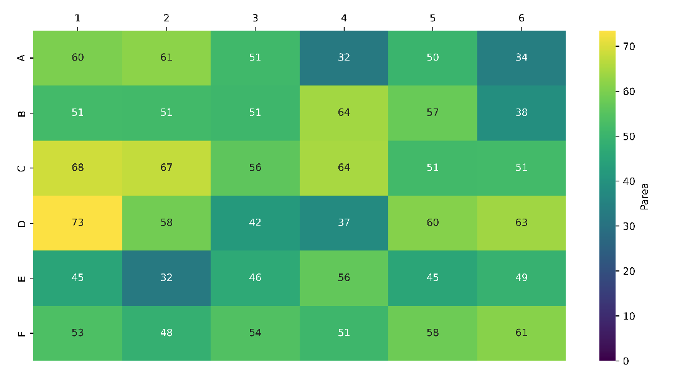
Figure 5: Heatmap showing 100% chemistry success rate and high average product peak areas for derivatisation of our partial degrader library with the recently developed one-pot azidation/CuAAC protocol.
Summary and Outlook
In our pursuit of efficiency and innovation in compound screening methodologies, we are proud to announce the development of a novel D2B protocol at Domainex. Our previously developed D2B protocol utilises amide coupling chemistry, enabling the rapid elaboration of amine libraries for biological screening. This approach has already demonstrated its efficacy in house for the synthesis and screening of potential novel degraders.
Building upon this foundation, we have now validated a novel azidation/CuAAC two-step one-pot protocol, offering a complementary pathway to access a distinct chemical space for rapid biological screening. Unlike traditional methods that rely on expensive azide libraries, our innovative approach circumvents this limitation, providing a cost-effective means to explore triazole linkages.
Looking ahead, we aim to validate further synthetic methodologies for use in D2B screening, such that this rapid approach to hit identification can access a wide range of drug-like chemical space.
References
C. E. Hendrick et al. ACS Med. Chem. Lett. 2022, 13, 1182–1190.
Z. Wang et al. Nat. Commun. 2023, 14, 8437.
R. P. Thomas et al. Chem. Sci. 2021, 12, 12098–12106.
P. Gehrtz et al. J. Med. Chem. 2022, 65, 10341–10356.